About vpCells
This interactive web atlas of intron-tagged proteins is a companion to the publication Pooled multicolor tagging for visualizing subcellular protein dynamics [Reicher*, Reiniš* et al].
We present a collection of fluorescence microscopy images of 1,158 tagged proteins in 4,576 isolated clonal cell lines (HAP1, HEK293T cells). It has been established as a part of our Visual Proteomics approach, described in more detail below and in full detail in the publication. Each protein of interest in this collection is endogenously tagged with either EGFP (frame 0 indicated in sgRNA details) or mScarlet (frame 1). Frozen clones are available upon request – in case you are interested in a particular protein, please reach out to vpcells@cemm.at. Please be aware that besides the protein of interest EGFP or mScarlet, other fluorophores are present in each clone as described below. We decided to include the entire dataset of over 75,000 images and not remove clones where the tagged proteins do not match the expected localization or where there is a possible mix of clones in the well.
Visual Proteomics
The subcellular localizations of proteins can be highly dynamic and change during many cellular processes or in response to different perturbations. Proteins can shuttle between compartments, localize to membrane-less organelles, bind to subcellular structures or form complexes. In different compartments or different environments, the same protein can carry out different functions. Therefore, knowing the subcellular localizations of proteins is essential for a complete understanding of cellular biology.
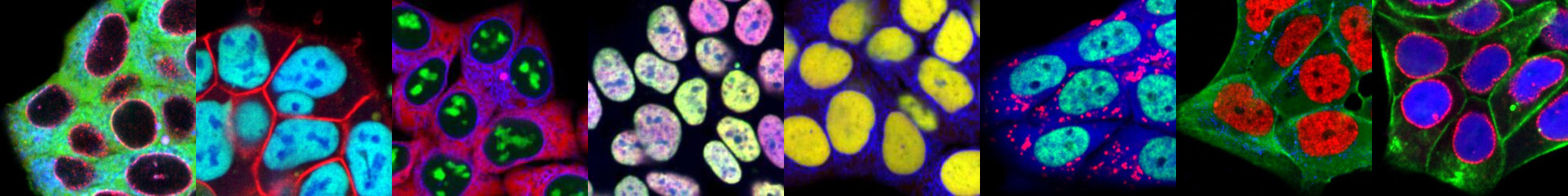
Examples of vpCells clones.
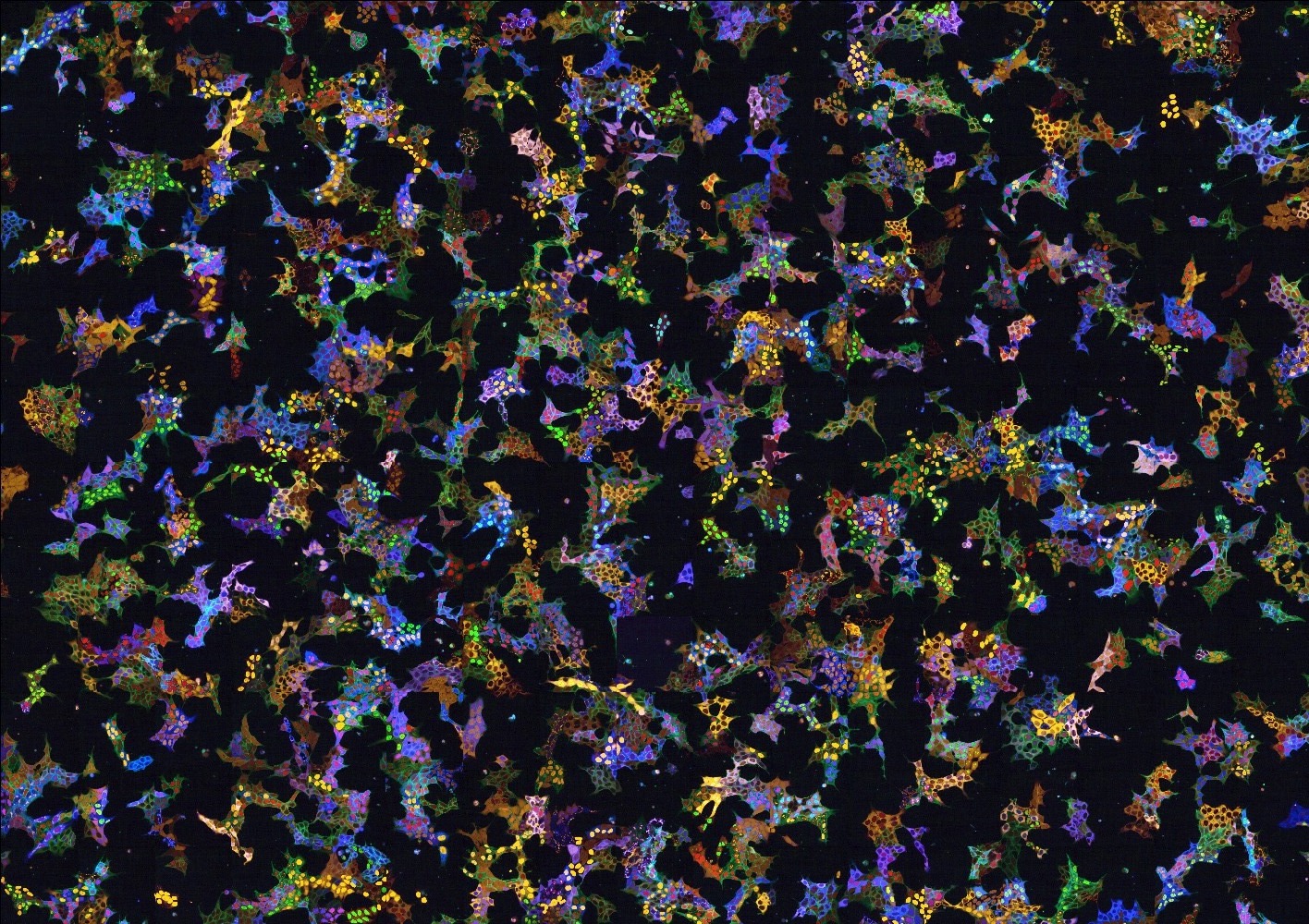
A vpCells pool (HAP1 cells).
Generation of vpCells and the clone collection
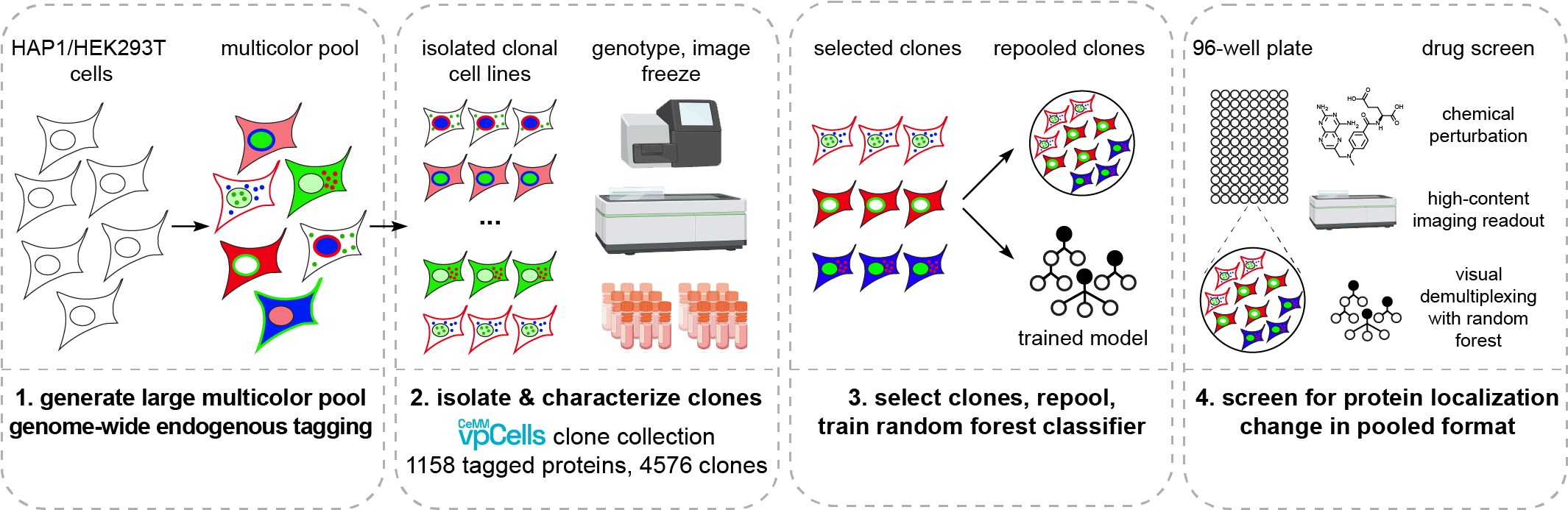
An overview of our Visual Proteomics approach.
To generate vpCells, we use a CRISPR-Cas9-based intron tagging strategy to generate cell pools expressing hundreds of endogenously tagged proteins. In brief, cells are transduced with an intron-targeting sgRNA library and a synthetic exon containing the coding sequence of EGFP is integrated at target sites. Fluorescent cells are sorted to obtain a cell pool, where a different protein is tagged in every cell, depending on the intron-targeting sgRNA expressed by that cell. This process is repeated and in every cell, a second protein is tagged with mScarlet in a second round of intron tagging. Then, the fluorescent protein mAmetrine localized to the plasma membrane and a far red fluorescent protein localizing to the nucleus are added. These additional channels are required in image analysis for segmentation and to further increase the complexity required to discriminate more clones in the pool.
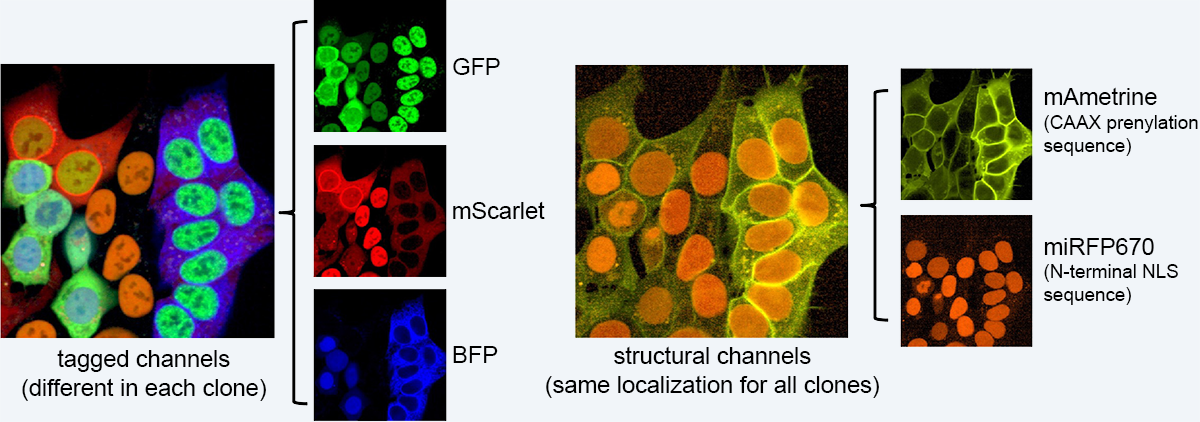
Overview of fluorophores in vpCells clones.
Finally, thousands of individual clones are isolated from the pools by single cell dilution. These isolated clonal cell lines are genotyped and imaged to train a computational model that is capable of recognizing the clones in the pool. With that computational model, vpCells can finally be used for monitoring changes in the subcellular localization and abundance of hundreds of proteins in a pooled format.
The detailed descriptions of sgRNA libraries we designed and used are available in CSV/Excel format in the Downloads section.
© 2022 - 2025 CeMM (v1.0.4)